Longest common subsequence
I pinched the idea for this animation from Ned Batchelder’s clever visualisation of Román Cortés’ brilliant CSS Homer Simpson. It depends on Javascript, CSS and a font containing upwards, leftwards and north west arrows. If it doesn’t work, my apologies. If you’d like to find out why I’m comparing humans with chimpanzees, read on!
Retracing our steps
Taking a brief step back, this article is the third of a series. In the first episode we posed a puzzle:
Starting with a list of runners ordered by finishing time, select a sublist of runners who are getting younger. What is the longest such sublist?
In the second episode we coded up a brute force solution which searched all possible sublists to find an optimal solution. Although the code was simple and succinct, its exponential complexity made it unsuitable for practical use.
In this episode we’ll discuss an elegant algorithm which solves our particular problem as a special case. On the way we’ll visit dynamic programming, Python decorators, version control and genetics.
An outline of the approach is:
- forget about races, runners and ages
- the essence of the problem is to determine the longest ordered subsequence found within a given sequence R
- which is a measure of how close R is to being ordered
- so, take a copy of R and sort it using our ordering criterion to form the sequence S
- the subsequences of R which are also subsequences of S are precisely the ordered subsequences of R
- so any longest subsequence common to both R and S is actually a longest ordered subsequence of R
- which is exactly what we’re looking for!
Copying and sorting finite sequences is straightforward (and efficient implementations can be found in any standard library), so the steps above reduce our problem to an application of a more general problem: how to find the longest common subsequence of two sequences.
Simple example
As a simple example I’ve highlighted “HMAN”, the longest subsequence common to both “HUMAN” and “CHIMPANZEE”
H U M A N
C H I M P A N Z E E
Let’s note up front that there may not be a unique LCS of two sequences (as a trivial example, “A” and “B” are both LCSes of “AB”, “BA”), which is why we’ll talk about a longest common subsequence rather than the longest common subsequence.
We can also rule out a simple divide and conquer scheme. We can’t just split both inputs in half and join together the LCSes of the beginnings and endings to form an answer. Consider finding the LCS of “123ABC”, “DEF123” for example — concatenating the LCSes of “123”, “DEF” and “ABC”, “123” gives the empty string, which is clearly wrong. (We’ll see later that a more sophisticated divide and conquer scheme does exist, but it comes at a cost.)
A recursive approach
An elegant recursive approach to solving this problem is based around the observation that the LCS of two sequences can be built from the LCSes of prefixes of these sequences. In algorithm speak, the longest common subsequence problem has optimal substructure.
def lcs(xs, ys):
'''Return a longest common subsequence of xs and ys.
Example
>>> lcs("HUMAN", "CHIMPANZEE")
['H', 'M', 'A', 'N']
'''
if xs and ys:
*xb, xe = xs
*yb, ye = ys
if xe == ye:
return lcs(xb, yb) + [xe]
else:
return max(lcs(xs, yb), lcs(xb, ys), key=len)
else:
return []
A formal explanation of why this works would be much longer than the code. I’ll sketch out the reasoning, but for a more rigorous approach I recommend Cormen, Leiserson, Rivest and Stein’s classic reference, “Introduction to Algorithms” (CLRS).
Essentially, if either input is empty, then the answer is just the empty sequence. Otherwise, split xs into its beginning, xb, and its final element xe; and similarly for ys. If the value xe equals ye, then lcs(xs, ys) must end with this value, and the remainder of lcs(xs, ys) must solve lcs(xb, yb). Otherwise, the longer of lcs(xs, yb) and lcs(xb, ys) must be a solution to lcs(xs, ys).
By the way, the assignment *xb, xe = xs is a nice example of Python 3.0’s extended unpacking syntax. In Python 2.6 and earlier you might write xb, xe = xs[:-1], xs[-1].
The recursion here is interesting. At each step, we compare two elements from the original inputs. Depending on the result, we must evaluate either one or two smaller LCSes.
Note also the call to max in the recursion. At this point, the algorithm chooses between two prefix pairs of the original inputs, (xs, yb) and (xb, ys), based on the length of their LCSes. Here’s the source of multiple solutions: max() returns the larger of its two arguments, and if lcs(xs, yb) and lcs(xb, ys) have equal lengths, I suspect the first will always be selected; but if we wanted all LCSes rather than any one LCS, we would have to continue down both routes.
The other important point to make here is that, in any case, both routes must be fully evaluated before a choice can be made. Loosely speaking, every additional step can double the workload. This is a problem!
Problems with the recursive solution
Work is needed to turn the recursive solution into a practical one. The key issue is that the recursive algorithm calculates the LCS of the same prefix pair multiple times. Let’s run a quick code experiment to show this. Starting with the version of lcs() shown above, we can adapt it to keep track of call counts1.
from collections import defaultdict
concat = ''.join
def lcs(xs, ys):
....
def count_lcs_calls(lcs):
'''Return a pair (lcs, calls)
Where:
lcs - a wrapped version of lcs, which counts up calls
calls - a dict mapping arg pairs to the number of times lcs
has been called with these args.
'''
calls = defaultdict(int)
def wrapped(xs, ys):
calls[(concat(xs), concat(ys))] += 1
return lcs(xs, ys)
return wrapped, calls
lcs, calls = count_lcs_calls(lcs)
If count_lcs_calls() reminds you of a Python decorator, good! That’s pretty much how it works. We can now call the newly decorated version of lcs and examine the calls dict to see what’s been going on.
With two completely different inputs, the recursive algorithm never passes the xe == ye test, and must always consider another pair of prefixes of the inputs.
>>> lcs('MAN', 'PIG')
[]
>>> print(*sorted((v, k) for k, v in calls.items()), sep='\n')
(1, ('', 'PIG'))
(1, ('M', 'PIG'))
(1, ('MA', 'PIG'))
(1, ('MAN', ''))
(1, ('MAN', 'P'))
(1, ('MAN', 'PI'))
(1, ('MAN', 'PIG'))
(2, ('MA', 'PI'))
(3, ('', 'PI'))
(3, ('M', 'PI'))
(3, ('MA', ''))
(3, ('MA', 'P'))
(6, ('', 'P'))
(6, ('M', ''))
(6, ('M', 'P'))
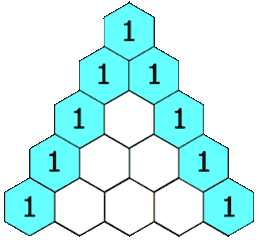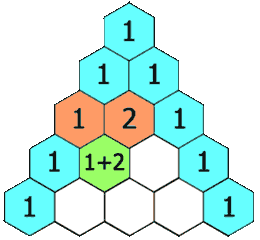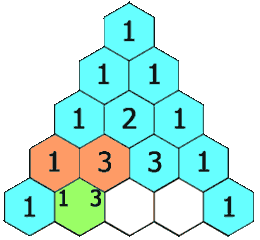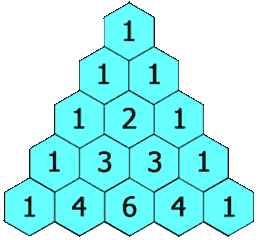
Here, we show the LCS of “MAN” and “PIG is the empty sequence. The next command prints the call counts to standard output, showing that e.g. we calculate lcs('MA', 'PI') twice and lcs('M', 'P') six times. It should be no surprise to see these call counts appearing in Pascal’s triangle.
At the other extreme, given identical inputs the recursion zips straight through the identical prefixes, and no call is repeated.
>>> calls.clear()
>>> lcs('CHIMP', 'CHIMP')
['C', 'H', 'I', 'M', 'P']
>>> print(*sorted((v, k) for k, v in calls.items()), sep='\n')
(1, ('', ''))
(1, ('C', 'C'))
(1, ('CH', 'CH'))
(1, ('CHI', 'CHI'))
(1, ('CHIM', 'CHIM'))
(1, ('CHIMP', 'CHIMP'))
Generally, though, we won’t be so lucky. By the time I interrupted a call to find the LCS of two short strands of DNA, there had already been a total of over 200 million recursive calls to the function, and we’d compared 'A' and 'G' alone over 24 million times. The algorithm, as presented, is of exponential complexity, hence suitable for only the briefest of inputs.
>>> lcs('ACCGGTCGAGTGCGCGGAAGCCGGCCGAA', 'GTCGTTCGGAATGCCGTTGCTCTGTAAA')
....
KeyboardInterrupt
>>> sum(calls.values())
227603392
>>> max(calls.values())
24853152
>>> import operator
>>> value = operator.itemgetter(1)
>>> mostcalled = list(sorted(calls.items(), key=value))[-5:]
>>> print(*mostcalled, sep='\n')
(('', 'GT'), 13182769)
(('A', 'GT'), 13182769)
(('A', 'G'), 24853152)
(('', 'G'), 24853152)
(('A', ''), 24853152)
Dynamic programming
We’ve already seen the LCS of two sequences can be built from the LCSes of prefixes of these subsequences; that is, an optimal solution to the problem can be built from optimal solutions to subproblems, a property known as optimal substructure. Our code experiment above shows that the subproblems repeat, possibly many times. This second property, of overlapping subproblems, means we have a classic example of a problem we can solve using dynamic programming. There are a couple of standard ways to progress:
- memoization
- converting from top-down to bottom-up
Let’s try these out.
Memoize
Memoization is a simple idea. Rather than repeatedly recalculate a function for the same input values, we cache the result of each call, keyed by the function call arguments. If, as in this case, full evaluation is expensive compared to the look-up, and we’ve got space for the cache, then the technique makes sense.
In Python, we can write an adaptor function which converts our original function into a memoized version of itself. Here’s just such an adaptor for functions which take an arbitrary number of positional parameters.
def memoize(fn):
'''Return a memoized version of the input function.
The returned function caches the results of previous calls.
Useful if a function call is expensive, and the function
is called repeatedly with the same arguments.
'''
cache = dict()
def wrapped(*v):
key = tuple(v) # tuples are hashable, and can be used as dict keys
if key not in cache:
cache[key] = fn(*v)
return cache[key]
return wrapped
We can use this to decorate our longest common subsequence function. I’ve also taken the opportunity to bypass some of the list splitting, creating an inner function which uses indices into the original xs and ys inputs.
def lcs(xs, ys):
'''Return the longest subsequence common to xs and ys.
Example
>>> lcs("HUMAN", "CHIMPANZEE")
['H', 'M', 'A', 'N']
'''
@memoize
def lcs_(i, j):
if i and j:
xe, ye = xs[i-1], ys[j-1]
if xe == ye:
return lcs_(i-1, j-1) + [xe]
else:
return max(lcs_(i, j-1), lcs_(i-1, j), key=len)
else:
return []
return lcs_(len(xs), len(ys))
This is a big improvement. If xs and ys have nx and ny elements respectively, then the values of i and j passed to the inner lcs_(i, j) function must fall into the inclusive ranges [0, nx], [0, ny], and the cache will grow to contain at most (nx + 1)×(ny + 1) elements. If the input sequences are of similar size, it’s easy to see this version of lcs operates in quadratic time and space.
>>> ss = lcs('ACCGGTCGAGTGCGCGGAAGCCGGCCGAA', 'GTCGTTCGGAATGCCGTTGCTCTGTAAA')
>>> ''.join(ss)
'GTCGTCGGAAGCCGGCCGAA'
Recursion limit
The memoized version of lcs() remembers previous results, but it’s still highly recursive, and that can cause problems. Try comparing a couple of longer synthesised strands of DNA.
>>> from random import choice
>>> def base(): return choice('ACGT')
>>> dna1 = [base() for _ in range(10000)]
>>> dna2 = [base() for _ in range(10000)]
>>> lcs(dna1, dna2)
....
RuntimeError: maximum recursion depth exceeded
We might try bumping up the recursion limit but this approach is neither clean nor portable. As the sys.setrecursionlimit() documentation says:
The highest possible limit is platform-dependent. A user may need to set the limit higher when she has a program that requires deep recursion and a platform that supports a higher limit. This should be done with care, because a too-high limit can lead to a crash.
We might also try using a different language or a different Python implementation, but instead let’s stick with CPython and explore a bottom-up approach.
Bottom up calculation
Rather than continue with our top-down recursive algorithm, we can employ another standard dynamic programming technique and transform the algorithm into one which works from the bottom up. Recall that, in the general case, to calculate lcs(xs, ys) we needed to solve three sub-problems, lcs(xb, yb), lcs(xb, ys), lcs(xs, yb), where xb is the result of removing the end element from xs, and similarly for yb.
*xb, xe = xs
*yb, ye = ys
if xe == ye:
return lcs(xb, yb) + [xe]
else:
return max(lcs(xs, yb), lcs(xb, ys), key=len)
These subproblems in turn depend on the LCSes of yet shorter prefix pairs of xs and ys. A bottom up approach reverses the direction and calculates the LCSes of all prefix pairs of xs and ys, from shortest to longest. We iterate through these prefix pairs filling a rectangular grid with the information needed to continue and to eventually reconstruct an LCS. Once again, the code is easier to read than an explanation.
from collections import defaultdict, namedtuple
from itertools import product
def lcs_grid(xs, ys):
'''Create a grid for longest common subsequence calculations.
Returns a grid where grid[(j, i)] is a pair (n, move) such that
- n is the length of the LCS of prefixes xs[:i], ys[:j]
- move is \, ^, <, or e, depending on whether the best move
to (j, i) was diagonal, downwards, or rightwards, or None.
Example:
T A R O T
A 0< 1\ 1< 1< 1<
R 0< 1^ 2\ 2< 2<
T 1\ 1< 2^ 2< 3\
'''
Cell = namedtuple('Cell', 'length move')
grid = defaultdict(lambda: Cell(0, 'e'))
sqs = product(enumerate(ys), enumerate(xs))
for (j, y), (i, x) in sqs:
if x == y:
cell = Cell(grid[(j-1, i-1)].length + 1, '\\')
else:
left = grid[(j, i-1)].length
over = grid[(j-1, i)].length
if left < over:
cell = Cell(over, '^')
else:
cell = Cell(left, '<')
grid[(j, i)] = cell
return grid
The implementation here makes a couple of unusual choices. It models a 2 dimensional grid of values using a dict keyed by (i, j) pairs, rather than a more conventional array representation. Furthermore, it uses a defaultdict, which returns a value even for keys which haven’t been filled in. I’ve used this data structure primarily to avoid special case code at the edges of the grid. For example, a grid cell can be accessed at position (j-1, i-1) even if i or j is zero, and that cell automatically takes the desired (0, 'e') value.
I’ve come across the suggestion that defaultdict’s should be converted into regular dict’s before being passed back to callers, which makes a lot of sense, but in this case I’m choosing to return a defaultdict since its default value property will be useful in subsequent calculations — to find an actual LCS for example.
Extracting an LCS from the grid
Cells in the LCS grid hold moves and LCS lengths. To extract an actual LCS, we need to retrace the moves. Here’s an iterative algorithm (a recursive one would also be possible) which does this.
def lcs(xs, ys):
'''Return a longest common subsequence of xs, ys.'''
# Create the LCS grid, then walk back from the bottom right corner
grid = lcs_grid(xs, ys)
i, j = len(xs) - 1, len(ys) - 1
lcs = list()
for move in iter(lambda: grid[(j, i)].move, 'e'):
if move == '\\':
lcs.append(xs[i])
i -= 1
j -= 1
elif move == '^':
j -= 1
elif move == '<':
i -= 1
lcs.reverse()
return lcs
If you haven’t come across the 2-argument version of iter, it’s a neat way to generate a sequence terminated by a sentinel value. In this case we move back through the grid, finishing when we hit an 'e' end move. Note that we only require the move fields of the grid cells to reconstruct an LCS. The length fields were used during calculation of the grid itself.
Animation
I’ve reproduced here the animation shown at the start of the article. It demonstrates the LCS grid being calculated, one cell at a time. The numbers in the cells correspond to the lengths of the LCSes ending at various prefix pairs of the inputs and the arrows indicate the moves: as an example, the 2 in the 4th row, 4th column shows that lcs("CHIM", "HUMA") has length 2, and the leftwards arrow shows the best previous LCS was lcs("CHIM", "HUM").
Once the grid has been filled, the red and orange arrows track back through it to find an LCS. The orange arrows indicate matches — elements of an LCS — and the red arrows indicate a route which threads through in the greatest possible number of these.
Performance
If the inputs are of size X, Y, then the grid will have X × Y entries each of which gets set in constant time. Recovering an LCS from the grid is a linear operation, so overall, this algorithm operates in O(XY) time and requires O(XY) space. If X and Y are similarly sized, we have a quadratic algorithm.
Practically speaking, we don’t bang into CPython’s recursion limit any more, but large inputs still put a strain on the machine. I tried running this new LCS calculation on a couple of long strands of DNA.
>>> len(dna1), len(dna2) (10000, 10000) >>> lcs(dna1, dna2)
To complete this calculation, we’ll need a grid with 10000 × 10000 entries, i.e. 100 million entries. So if each grid entry occupies 10 bytes, we’re looking at roughly a gigabyte — a sizable chunk even for today’s computers. And if each step takes 100 nanoseconds, we’d be looking at a run-time of 10 seconds, if we don’t run out of memory first that is.
This back of an envelope calculation turned out to be way off. Running the code snippet above caused my system to thrash. I killed it after 10 minutes, at which point the Python process alone was using over 1GB of memory.
At this point the choice of a dict (a hashtable) very definitely becomes questionable. Actually, our choice of Python becomes questionable. When you have a quadratic algorithm with large inputs, the space and time constant factors both become critical.
We have a number of options:
- find a leaner algorithm.
- find a bigger computer.
- treat our Python implementation as a prototype. Code up a high performance version in a language which gives better control over the machine.
Then there’s option 2a, to find a way of distributing the job over a number of computers. There’s also an option 3a, to tune up our Python implementation.
Reducing the space for LCS lengths
When you’ve run out of main memory, any estimate of runtime based on big-O analysis becomes useless. The system either crashes or thrashes, paging in virtual memory. By contrast, if we can reduce the memory required by a program, the time analysis should still hold — we never have to page in more time.
Once you’ve watched the dynamic programming solution a few times, you’ll realise that the LCS lengths (the numbers in the grid) are computed row by row, with each row only depending on the row above. If we just wanted to calculate the LCS length of two sequences, we could write:
import itertools
def lcs_length(xs, ys):
'''Return the length of the LCS of xs and ys.
Example:
>>> lcs_length("HUMAN", "CHIMPANZEE")
4
'''
ny = len(ys)
curr = list(itertools.repeat(0, 1 + ny))
for x in xs:
prev = list(curr)
for i, y in enumerate(ys):
if x == y:
curr[i+1] = prev[i] + 1
else:
curr[i+1] = max(curr[i], prev[i+1])
return curr[ny]
This LCS length algorithm requires just a linear amount of space for the current and previous rows in the grid (in fact we could halve this space again, since each the LCS calculation for each cell in the grid only depends on the contents of the previous ny + 2 cells, but the code is a little more subtle).
To calculate an actual LCS, though, we would still need a full grid of moves for the final backtracking stage of the algorithm. Clearly we can compact this grid, since each cell contains one of four moves (↑, ←, ↖ e) and can therefore be be represented in just 2 bits. Despite this, the grid would still occupy a quadratic amount of space.
Fortunately, an ingenious hybrid algorithm by Professor Dan Hirschberg requires just a linear amount of space.
Hirschberg’s Linear Space Algorithm
In a 1975 paper titled “A linear space algorithm for computing maximal common subsequences”, D.S. Hirschberg describes a technique for computing an LCS using space proportional to the length of the inputs. The algorithm combines the dynamic programming technique discussed in this article with a classic divide and conquer approach.
If you search for this paper, you’ll soon find a copy of the original online. It’s just three pages long and very accessible. The key idea is to split one of the input sequences in two.
>>> mid = len(xs)//2 >>> xb, xe = xs[:mid], xs[mid:]
We then calculate LCS lengths for the pair (xb, ys) and the pair (reversed(xe), reversed(ys)). If we find a y-position, k, which maximises the sums of these forward and backwards LCS lengths, then we have a suitable position to split ys. Thus our answer reduces to finding the answer for two smaller subsequences.
>>> yb, ye = ys[:k], ys[k:] >>> lcs_xs_ys = lcs(xb, yb) + lcs(xe, ye)

The picture shows the first stage of this algorithm for the input pair (PYTHON, PONY).
- split PYTHON evenly into PYT, HON
- working forwards from the top left, use
lcs_length('PYT', 'PONY)'to fill the T row of the grid. - working backwards from the bottom right, use
lcs_length('NOH', 'YNOP)'to fill the H row of the grid - find the column which maximises the sum of lcs lengths on the T and H rows, adding each T-item to the H-item below and one to the right. In this case, the sums are + 2, 1 + 2, 1 + 1, 1 + 0, 2 +, the maximum value of 3 is at 1 + 2.
- split PONY at this column.
- thus
lcs('PYTHON', 'PONY')is equal tolcs('PYT', 'P') + lcs('HON', 'ONY')
Hirschberg proves the correctness of this algorithm and goes on to prove that it has quadratic performance but requires just a linear amount of space.
The code looks something like the following.
import itertools
def lcs_lens(xs, ys):
curr = list(itertools.repeat(0, 1 + len(ys)))
for x in xs:
prev = list(curr)
for i, y in enumerate(ys):
if x == y:
curr[i + 1] = prev[i] + 1
else:
curr[i + 1] = max(curr[i], prev[i + 1])
return curr
def lcs(xs, ys):
nx, ny = len(xs), len(ys)
if nx == 0:
return []
elif nx == 1:
return [xs[0]] if xs[0] in ys else []
else:
i = nx // 2
xb, xe = xs[:i], xs[i:]
ll_b = lcs_lens(xb, ys)
ll_e = lcs_lens(xe[::-1], ys[::-1])
_, k = max((ll_b[j] + ll_e[ny - j], j)
for j in range(ny + 1))
yb, ye = ys[:k], ys[k:]
return lcs(xb, yb) + lcs(xe, ye)
I say “something like” because this code is unpolished and inefficient. Although the space requirement is proportional to the input sizes, it continually creates new list slices in order to get the job done. It may well be capable of handling large inputs in a using a limited amount of memory, but it takes far too long doing so.
On a more positive note, at least this implementation passes some simple tests.
def test():
concat = ''.join
assert concat(lcs('', '')) == ''
assert concat(lcs('a', '')) == ''
assert concat(lcs('', 'b')) == ''
assert concat(lcs('abc', 'abc')) == 'abc'
assert concat(lcs('abcd', 'obce')) == 'bc'
assert concat(lcs('abc', 'ab')) == 'ab'
assert concat(lcs('abc', 'bc')) == 'bc'
assert concat(lcs('abcde', 'zbodf')) == 'bd'
assert concat(lcs('aa','aaaa')) == 'aa'
assert concat(lcs('GTCGTTCGGAATGCCGTTGCTCTGTAAA',
'ACCGGTCGAGTGCGCGGAAGCCGGCCGAA')
) == 'GTCGTCGGAAGCCGGCCGAA'
This is an achievment in itself: if you examine the implementation, you’ll see plenty of space for off-by-one errors to creep in. This is about as far as we’ll get with Python, but a working implementation is a great resource when we switch to a more efficient language.
Applied to the New York Marathon Results
Remember our original problem? At last we’re ready to take it on.
In 2008, 38096 runners completed the world’s largest running event, the New York City marathon. A full set of results is available online, one page at a time, but it’s not too hard to scrape them into a single file for convenient offline processing.
Runners’ ages are supplied to the nearest year. To find a longest sublist of the results comprising runners who aren’t getting any older, we sort the results in reverse age order, then run our LCS algorithm against the original results list. Note that we use age alone for comparison: so, for example, GAUTI HOSKULDSSON (a 47 year old male who finished in 295th place with a time of 2:47:00) equals PHILIPPE LETESSIER (also 47, male, 298th in 2:47:14).
To find a longest sublist of runners who are getting strictly younger, we must uniquify the youth-ordered results before applying the LCS algorithm.
Here’s a sketch of how we’d do it in Python. I’ve omitted details of the AgingRunner class; all that matters is that it has an age field, which is used for both ordering and equality comparisons.
class AgingRunner:
'''Class to represent runners keyed by age.
'''
....
def __lt__(self, other):
'''Return True if this runner is *older* than the other.'''
return self.age > other.age
def __eq__(self, other):
'''Return True if this runner is the same age as the other.'''
return self.age == other.age
def uniq(xs):
import itertools, operator
return map(operator.itemgetter(0), itertools.groupby(xs))
runners = read_in_results()
srunners = list(sorted(runners))
not_getting_older = lcs(runners, srunners)
urunners = list(uniq(srunners))
getting_younger = lcs(runners, urunners)
Sadly this really is just a sketch. Even though our space efficient Python LCS implementation runs comfortably in main memory on a modern computer, it just isn’t quick enough. Hirschberg’s paper (published way back in 1975) says:
We present an algorithm which will solve this problem in quadratic time and in linear space. For example, assuming coefficients of 2 microseconds and 10 bytes […] for strings of length 10,000 we would require a little over 3 minutes and 100K bytes.
If we suppose a modern computer runs 1000 times faster, but that our naive Python implementation is 100 times slower than native code, for input sizes of 38096 we’d expect a run time of around 6 minutes. Sadly this estimate turns out to be optimistic. It took me almost an hour and a half to run this program on a computer with a 2GHZ Intel Core duo processor.
C++ excels at high performance sequence crunching. Rather than try and tune up my Python program I used it as a reference implementation for a C++ one. The C++ version processed the full set of results in a respectable 18 seconds, giving a coefficient of around 12 nanoseconds — much closer to the figure I’d expect from a machine with a 2.33 GHz processor.
I found the longest sequence of runners who weren’t getting any older had length 1724. I also found a sequence of 60 runners who were getting younger, covering every age between 80 and 18, with the exceptions of 79, 77, 76.
24319, CHABANE KETFI, 80, M, 4:37:04 31293, GRACCO VARAGNOLO, 78, M, 5:07:42 32576, JOSEPH PASCARELLA, 75, M, 5:17:32 32931, THEODORE ROGERS, 74, M, 5:20:42 33351, GOODMAN ESPY, 73, M, 5:24:17 33982, MANUEL GUARIN, 72, M, 5:30:06 34646, GIORGIO LUDWIG, 71, M, 5:37:43 35014, PIETRO ROSSI, 70, M, 5:42:00 35052, MARCELLA HSIUNG, 69, F, 5:42:41 35717, LUIS BECERRA, 68, M, 5:52:55 35841, SHIZUKO HAZEYAMA, 67, F, 5:55:25 36237, MONIKA LUDWIG, 66, F, 6:03:30 36248, PETER GELTNER, 65, M, 6:03:49 36427, SERGE GIROUD, 64, M, 6:08:44 36462, HIDEMI HARA, 63, F, 6:09:58 36483, KIMIE NISHIOKA, 62, F, 6:10:24 36486, ERNEST YONG, 61, M, 6:10:33 36530, FRANCO ZUCCHINI, 60, M, 6:11:39 36629, FRANCESCO CERETTI, 59, M, 6:14:18 36646, MASAHARU AKAZAWA, 58, M, 6:14:39 36680, LOUISE SEELEY, 57, F, 6:15:31 36706, BETH EIDEMILLER, 56, F, 6:16:22 36720, DANIELA CARACENI, 55, F, 6:16:51 36796, GUSTAVO CASTILLO SOSA, 54, M, 6:18:53 36836, GAIL COOPER, 53, F, 6:19:49 36850, WAYNE DOLNICK, 52, M, 6:19:56 36869, ANNALEA BIANCHI, 51, F, 6:20:18 36914, PATRICIA DRISCOLL, 50, F, 6:21:59 ....
Incidentally, CHABANE KETFI was comfortably the fastest of 17 octogenarian finishers, one of whom, PETER HARANGOZO, 87, was also the oldest finisher.
Hirschberg’s linear space algorithm in C++
Here’s a C++ implementation based on the reference Python implementation. It should work on any container type which supports forward and reverse iteration. It’s similar to the Python implementation, but note:
- rather than copy slices of the original sequences, it passes around iterators and reverse iterators into these sequences
- rather than recursively accumulating sums of sub-LCS results, it ticks off items in a members array,
xs_in_lcs
/*
C++ implementation of
"A linear space algorithm for computing maximal common subsequences"
D. S. Hirschberg
http://portal.acm.org/citation.cfm?id=360861
See also: http://wordaligned.org/articles/longest-common-subsquence.html
*/
#include <algorithm>
#include <iterator>
#include <vector>
typedef std::vector<int> lengths;
/*
The "members" type is used as a sparse set for LCS calculations.
Given a sequence, xs, and members, m, then
if m[i] is true, xs[i] is in the LCS.
*/
typedef std::vector<bool> members;
/*
Fill the LCS sequence from the members of a sequence, xs
x - an iterator into the sequence xs
xs_in_lcs - members of xs
lcs - an output results iterator
*/
template <typename it, typename ot>
void set_lcs(it x, members const & xs_in_lcs, ot lcs)
{
for (members::const_iterator xs_in = xs_in_lcs.begin();
xs_in != xs_in_lcs.end(); ++xs_in, ++x)
{
if (*xs_in)
{
*lcs++ = *x;
}
}
}
/*
Calculate LCS row lengths given iterator ranges into two sequences.
On completion, `lens` holds LCS lengths in the final row.
*/
template <typename it>
void lcs_lens(it xlo, it xhi, it ylo, it yhi, lengths & lens)
{
// Two rows of workspace.
// Careful! We need the 1 for the leftmost column.
lengths curr(1 + distance(ylo, yhi), 0);
lengths prev(curr);
for (it x = xlo; x != xhi; ++x)
{
swap(prev, curr);
int i = 0;
for (it y = ylo; y != yhi; ++y, ++i)
{
curr[i + 1] = *x == *y
? prev[i] + 1
: std::max(curr[i], prev[i + 1]);
}
}
swap(lens, curr);
}
/*
Recursive LCS calculation.
See Hirschberg for the theory!
This is a divide and conquer algorithm.
In the recursive case, we split the xrange in two.
Then, by calculating lengths of LCSes from the start and end
corners of the [xlo, xhi] x [ylo, yhi] grid, we determine where
the yrange should be split.
xo is the origin (element 0) of the xs sequence
xlo, xhi is the range of xs being processed
ylo, yhi is the range of ys being processed
Parameter xs_in_lcs holds the members of xs in the LCS.
*/
template <typename it>
void
calculate_lcs(it xo, it xlo, it xhi, it ylo, it yhi, members & xs_in_lcs)
{
unsigned const nx = distance(xlo, xhi);
if (nx == 0)
{
// empty range. all done
}
else if (nx == 1)
{
// single item in x range.
// If it's in the yrange, mark its position in the LCS
xs_in_lcs[distance(xo, xlo)] = find(ylo, yhi, *xlo) != yhi;
}
else
{
// split the xrange
it xmid = xlo + nx / 2;
// Find LCS lengths at xmid, working from both ends of the range
lengths ll_b, ll_e;
std::reverse_iterator<it> hix(xhi), midx(xmid), hiy(yhi), loy(ylo);
lcs_lens(xlo, xmid, ylo, yhi, ll_b);
lcs_lens(hix, midx, hiy, loy, ll_e);
// Find the optimal place to split the y range
lengths::const_reverse_iterator e = ll_e.rbegin();
int lmax = -1;
it y = ylo, ymid = ylo;
for (lengths::const_iterator b = ll_b.begin();
b != ll_b.end(); ++b, ++e)
{
if (*b + *e > lmax)
{
lmax = *b + *e;
ymid = y;
}
// Care needed here!
// ll_b and ll_e contain one more value than the range [ylo, yhi)
// As b and e range over dereferenceable values of ll_b and ll_e,
// y ranges over iterator positions [ylo, yhi] _including_ yhi.
// That's fine, y is used to split [ylo, yhi), we do not
// dereference it. However, y cannot go beyond ++yhi.
if (y != yhi)
{
++y;
}
}
// Split the range and recurse
calculate_lcs(xo, xlo, xmid, ylo, ymid, xs_in_lcs);
calculate_lcs(xo, xmid, xhi, ymid, yhi, xs_in_lcs);
}
}
// Calculate an LCS of (xs, ys), returning the result in an_lcs.
template <typename seq>
void lcs(seq const & xs, seq const & ys, seq & an_lcs)
{
members xs_in_lcs(xs.size(), false);
calculate_lcs(xs.begin(), xs.begin(), xs.end(),
ys.begin(), ys.end(), xs_in_lcs);
set_lcs(xs.begin(), xs_in_lcs, back_inserter(an_lcs));
}
Applications: edit distance
You may be thinking this is all of academic interest only. Who really cares about the longest common subsequence problem? Well, anyone who uses version control does. The longest common subsequence problem is closely related to the more general edit distance problem, which is all about finding the smallest number of point mutations needed to change one sequence into another. For example, if we allow just two such mutations:
- insert an element
- delete an element
then the number of mutations needed to convert a xs into ys is the sum of the lengths of xs and ys minus twice the length of their LCS. Or in Python:
>>> edit_distance(xs, ys) == len(xs) + len(ys) - 2 * lcs(xs, ys) True
For example, we can change “HUMAN” into “CHIMPANZEE” using the LCS, “HMAN” as follows: delete the U from HUMAN, to give HMAN, then insert C, I, P, Z, E, E giving CHIMPANZEE.
In general:
- find an LCS of
xsandys. Let’s say this LCS has lengthL. - strike out all elements of
xswhich are not in this LCS, a total oflen(xs) - Lmoves - now insert all the elements of
yswhich are missing from the LCS, anotherlen(ys) - Lmoves.
Thus the edit distance is at most len(xs) + len(ys) - 2 * L. It remains to show there can be no shorter edit distance. The same argument works well enough in reverse. Given a minimal edit sequence, note we do not delete anything we already inserted, otherwise we could remove this insert-delete pair to form a shorter edit sequence. Thus we can sort this edit sequence so the deletions preceed the insertions to form another minimal edit sequence. . Now process all the deletions, leaving a subsequence of xs. Since we can now get from this subsequence to ys using insertions alone, it must also be a subsequence of ys.
More general variants of the edit distance problem allow substitution and transposition mutations, and may assign weights to each type of mutation. In all cases, similar algorithms can be used to the dynamic programming algorithm shown in this example, but improvements can be made if we know the inputs are similar — as in the case of genetic mutations, for example.
Applications: genetic mutations
I’ve already mentioned Cormen, Leiserson, Rivest and Stein’s classic reference, “Introduction to Algorithms” (CLRS for short). This book introduces the longest common subsequence problem as follows:
In biological applications, we often want to compare the DNA of two (or more) different organisms. […] For example, the DNA of one organism may be S1 = ACCGGTCGAGTGCGCGGAAGCCGGCCGAA while the DNA of another organism may be S2 = GTCGTTCGGAATGCCGTTGCTCTGTAAA. One goal of comparing two strands of DNA is to determine how “similar” the two strands are, as some measure of how cosely related the two organisms are.
And, CLRS goes on to say, the longest common subsequence of S1 and S2 is one such measure of similarity. In this case, the longest common subsequence, S3 would be GTCGTCGGAAGCCGGCCGAA
Genetic comparision is an important application of sequence matching. As already mentioned, it’s also a special case, which means we can can do rather better than the general purpose algorithm presented above. For one thing, the alphabet of bases, Adenine, Cytosine, Guanine, Thymine has just 4 elements. More importantly, the evolution of life on this planet is based around DNA replication. Over 400 million years, glitches in DNA replication lead to genetic mutations, leading in turn to a variety of life forms. But, for example, human and chimpanzee DNA are at least 96% similar. If we’re comparing two human genomes, a recent article entitled Human genomes as email attachments points out:
The key insight is to consider that any two human genomes are more than 99% identical.
So the random A, C, G, T sequences I used earlier this article were highly non-representative.
When the edit distance, D, between two sequences is small compared to the lengths of these sequences, more specialised algorithms can calculate this distance in O(ND) complexity and linear space. I’m not going to discuss these algorithms here, but they’re elegant and accessible. See a 1986 paper “An O(ND) difference algorithm and its variations” by Eugene W. Myers for more details.
Applications: version control
Similar considerations apply to a version controlled file. Generally the distance between two versions of a file is small compared to the size of the file itself; the Unix diff and patch tools exploit this, as do the difference engines embedded in version control systems. Here’s a comment in Subversion’s differencing engine.
/* * Calculate the Longest Common Subsequence between two datasources. * This function is what makes the diff code tick. * * The LCS algorithm implemented here is described by Sun Wu, * Udi Manber and Gene Meyers in "An O(NP) Sequence Comparison Algorithm" * */
(I’m fairly sure they mean “Myers”, and P is the the number of deletions in an edit sequence. You can easily find a copy of the referenced paper, which walks through a beautiful greedy algorithm and beats Myers’ earlier O(ND) algorithm by a factor of two, or better.)
Conclusions
The longest common subsequence problem and the more general edit distance problem have been — and continue to be — extensively studied. A classic dynamic programming solution to this problem is based on an elegant recursive algorithm, though by the time we’ve created an efficient implementation some of that elegance has been hidden. And efficiency, here, is an important consideration, since the general purpose algorithm presented in this article is of quadratic complexity.
A Python implementation is quick to develop and flexible enough to experiment with. With some effort we can control its space requirements. But a C++ implementation comfortably outperforms Python by an order of magnitude or more, and for large inputs, compiled native code is required. Does this make Python suitable only for toy projects, short scripts and blog posts? No, but I’ll save a more complete answer for another time!
In the case where the input sequences are similar, more efficient greedy algorithms exist. These algorithms form the backbone of the venerable Unix diff tool and of modern version control systems.
Next time
Let’s not forget that we really wanted to solve the longest ordered subsequence problem, not the longest common subsequence problem. In the next episode we’ll explore a beautiful — and efficient — longest ordered subsequence algorithm.
Thanks
My thanks to Soeren Meyer-Eppler for reporting a tricky bug in the C++ LCS implementation. I should have run more tests using -D_GLIBCXX_DEBUG -D_GLIBCXX_DEBUG_PEDANTIC!
Python 3 Notes
Note 1. The results served up from the official New York Marathon site were encoded using charset=iso-8859-1, and I maintained this encoding for my local copy of these results. In Python 3.0, you can explicitly specify an encoding when opening a file in text mode.
>>> fp = open('nyc-results.txt', encoding='iso-8859-1')
From the documentation:
encoding is the name of the encoding used to decode or encode the file. This should only be used in text mode. The default encoding is platform dependent, but any encoding supported by Python can be passed. See the codecs module for the list of supported encodings.
Somewhere along the line a case conversion algorithm must have failed to get the details right. Here are the finishers with more than one non-ASCII character in their names:
- 3275, TORBJöRN SKJöLDERFORS, 49, M, 3:22:46
- 4141, GöRAN HäRSTEDT, 43, M, 3:27:40
- 4510, JüRG MüLLER, 49, M, 3:29:15
- 6763, ØYVIND SCHAGE FøRDE, 35, M, 3:39:16
- 8832, ØYSTEIN ØKLAND, 55, M, 3:47:19
- 11402, THOMAS KRäHENBüHL, 31, M, 3:55:22
- 13582, LáSZLó SZABó, 50, M, 4:00:55
- 18186, ØYVIND BøRRESEN, 52, M, 4:16:31
- 23727, FERNANDO SEBASTIáN LóPEZ, 34, M, 4:34:50
- 28315, KOLBRúN BRYNJA EGILSDóTTIR, 46, F, 4:52:29
- 33457, JAN KRISTIAN RøDSTøL, 37, M, 5:25:12
Note 2. I’ve already pointed out Python 3.0’s extended unpacking syntax. Here’s another example:
>>> car, *cdr = items
Note 3. You may well have spotted the truncating integer division used in the divide and conquer algorithm:
>>> i = nx // 2
Without the double slash, i would become a floating point number, and using it as slice index would raise a TypeError. The // floor division operator is available in Python 2.6 and earlier and I recommend using it for all C-style truncating integer divisions. Here’s the full history.
Note 4. To order and compare runners by age for the preliminary sort() and the subsequent lcs() calls, I added __lt__ and __eq__ methods to the AgingRunner class. In Python 2.6 the __cmp__ method could have been used instead of these rich comparison methods, but this is no longer supported in 3.0. A recipe by Raymond Hettinger demonstrates a class decorator which automatically creates all the rich comparisons methods from any subset of them (e.g. __gt__, __ge__, __le__ can all be sensibly derived from __lt__).
1. Note that the original version of lcs() works for any pair of sequences with equality comparable elements. The newly adapted version will only work for strings and byte arrays. This isn’t really a problem, since we’re just using it to run a quick code experiment.